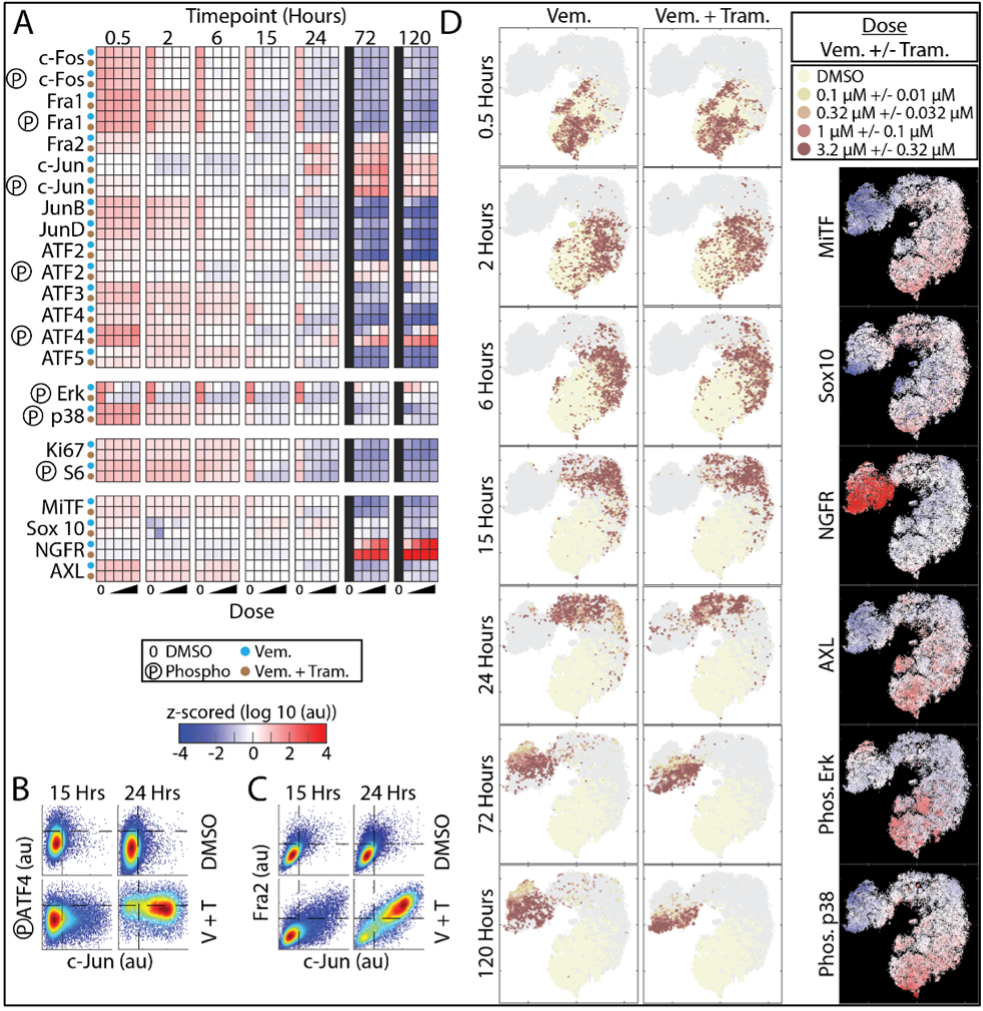
The overall goal of this project is to develop efficient computational tools to infer dynamic cell signaling networks as well as tumor-immune and immune-immune networks of interactions that play a key role in cancer progression and response to therapy. Recent discoveries from single-cell studies have shown that populations of cells (even those that are genetically identical) do not respond similarly when exposed to environmental perturbations such as drugs, growth factors or immune cytokines. Such phenotypic heterogeneity is a common cause of the suboptimal efficacy of many cancer treatments. For example, they complicate cancer therapies by giving rise to heterogeneous subpopulations of drug-tolerant cells. Despite advances in our understanding of phenotypic variability, the mechanisms by which cells integrate dose- and time-dependent inputs from upstream pathways or interactions with other cell types (e.g., stromal and immune cells) to direct heterogeneous decisions are still poorly understood. Without deeper mechanistic insights, promising therapies that fail to account for cell-to-cell variability will continue to fall short of achieving the desired therapeutic outcome. Our goal in this project is, therefore, to gain a predictive and dynamic understanding of the mechanisms by which signaling networks process inputs initiated by cancer drugs and the immune microenvironment to direct heterogeneous cell fate decisions. We will integrate new experimental systems that enable us to rapidly collect highly multiplexed single-cell data across a variety of cell types, drug perturbations, doses, and timepoints with innovative mathematical tools for the analysis of complex networked data and interconnected systems, to identify signaling network dynamics in heterogenous cell populations during different perturbed conditions. We will utilize these dynamics to predict diverse cell response trajectories and identify conditions where a desired therapeutic outcome (e.g., tumor cell death) would be achieved. Our interdisciplinary team collaborate with Fallahi-Sichani Lab and Dolatshahi Lab in the Biomedical Engineering department in UVA.
